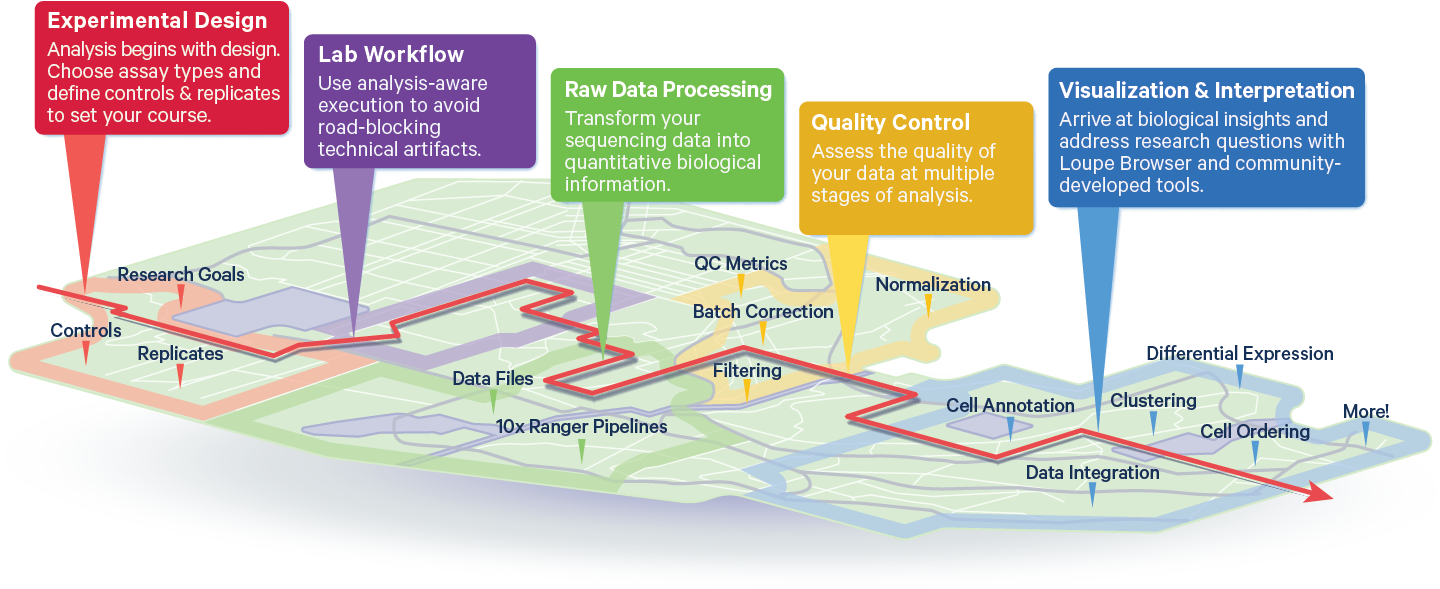
10x Genomics Analysis Journey
Introductions
Integrating Single Cell and Visium Spatial Gene Expression data
Integrating Single Cell and Visium Spatial Gene Expression data
Single cell and Visium gene expression data can be combined to elucidate spatiality in single cell data and improve resolution in Visium data.
Single cell and Visium gene expression data can be combined to elucidate spatiality in single cell data and improve resolution in Visium data.
Batch Effect Correction
Batch Effect Correction
Batch effects come from technical variation across samples. This can often be prevented with good experimental design. When it cannot, there are computational approaches that can help.
Batch effects come from technical variation across samples. This can often be prevented with good experimental design. When it cannot, there are computational approaches that can help.
Web Resources for Cell Type Annotation
Web Resources for Cell Type Annotation
The advantage of visualizing cell populations at single-cell resolutions has introduced us to the challenges of annotating cells. A growing number of annotation databases and tools are available to aid us in the process. In this article, we provide general guidance for a selected few to assist you with your research.
The advantage of visualizing cell populations at single-cell resolutions has introduced us to the challenges of annotating cells. A growing number of annotation databases and tools are available to aid us in the process. In this article, we provide general guidance for a selected few to assist you with your research.
Bioinformatics Tools for Sample Demultiplexing
Bioinformatics Tools for Sample Demultiplexing
Sample multiplexing can minimize batch effects, facilitate multiplet identification, lower experiment costs, and make large-scale sample operations practical. In this article, we introduce main methods along with the bioinformatics tools for sample demultiplexing.
Sample multiplexing can minimize batch effects, facilitate multiplet identification, lower experiment costs, and make large-scale sample operations practical. In this article, we introduce main methods along with the bioinformatics tools for sample demultiplexing.
Tutorials
Correcting Batch Effects in Visium Data
Correcting Batch Effects in Visium Data
Guided, step-by-step tutorial in R for correcting batch effects between two public 10x Genomics Visium datasets (mouse brain) using the Harmony algorithm, then importing corrected projections and clusters into the Loupe Browser.
Guided, step-by-step tutorial in R for correcting batch effects between two public 10x Genomics Visium datasets (mouse brain) using the Harmony algorithm, then importing corrected projections and clusters into the Loupe Browser.
Navigating 10x Genomics Barcoded BAM Files
Navigating 10x Genomics Barcoded BAM Files
The BAM file produced by the Ranger pipelines include standard SAM/BAM flags and several custom flags that can be used to mine information about alignment, annotation, counting, etc. This tutorial walks users through the process of identifying records in the BAM file that contribute to UMI counting.
The BAM file produced by the Ranger pipelines include standard SAM/BAM flags and several custom flags that can be used to mine information about alignment, annotation, counting, etc. This tutorial walks users through the process of identifying records in the BAM file that contribute to UMI counting.
Testing Microscope Image Compatibility with Space Ranger
Testing Microscope Image Compatibility with Space Ranger
Guides users through testing Loupe Browser and Space Ranger compatibility with images of various formats. Illustrates conversion to compatible TIFF in Fiji using multichannel CZI and multi-hue SVS images.
Guides users through testing Loupe Browser and Space Ranger compatibility with images of various formats. Illustrates conversion to compatible TIFF in Fiji using multichannel CZI and multi-hue SVS images.
Trajectory Analysis using 10x Genomics Single Cell Gene Expression Data
Trajectory Analysis using 10x Genomics Single Cell Gene Expression Data
This tutorial provides users with the instructions to import results obtained with Cell Ranger and Loupe Browser into community-developed tools for RNA velocity analysis. This analysis can be used to reconstruct the dynamic processes that cells undergo as part of their true biological nature.
This tutorial provides users with the instructions to import results obtained with Cell Ranger and Loupe Browser into community-developed tools for RNA velocity analysis. This analysis can be used to reconstruct the dynamic processes that cells undergo as part of their true biological nature.
Batch Effect Correction in Chromium Single Cell ATAC Data
Batch Effect Correction in Chromium Single Cell ATAC Data
A step-by-step tutorial for batch effect correction in two 10x Genomics Single Cell ATAC datasets using the Harmony algorithm in R. The corrected projection was then imported into the Loupe Browser.
A step-by-step tutorial for batch effect correction in two 10x Genomics Single Cell ATAC datasets using the Harmony algorithm in R. The corrected projection was then imported into the Loupe Browser.
Tag Assignment of 10x Genomics CellPlex Data Using Seurat’s HTODemux Function
Tag Assignment of 10x Genomics CellPlex Data Using Seurat’s HTODemux Function
A step-by-step tutorial for using Seurat’s HTODemux function to perform custom tag assignment of 10x Genomics CellPlex data. The output tag assignments can be loaded back into Cell Ranger to rerun the primary analysis.
A step-by-step tutorial for using Seurat’s HTODemux function to perform custom tag assignment of 10x Genomics CellPlex data. The output tag assignments can be loaded back into Cell Ranger to rerun the primary analysis.
Background Removal Guidance for Single Cell Gene Expression Datasets Using CellBender
Background Removal Guidance for Single Cell Gene Expression Datasets Using CellBender
A guided tutorial for removing background in Single Cell Gene Expression data using the community developed tool CellBender. The ‘cleaned’ dataset is further validated by cell type annotation and visualization.
A guided tutorial for removing background in Single Cell Gene Expression data using the community developed tool CellBender. The ‘cleaned’ dataset is further validated by cell type annotation and visualization.
Integrating 10x Visium and Chromium data with R
Integrating 10x Visium and Chromium data with R
A step-by-step guide for using spacexr to deconvolve Visium spatial transcriptomic spot cell types using a Chromium single cell reference.
A step-by-step guide for using spacexr to deconvolve Visium spatial transcriptomic spot cell types using a Chromium single cell reference.
Informatics Blogs
Continuing Your Journey After Running Cell Ranger
Continuing Your Journey After Running Cell Ranger
Guidance on next steps after running Cell Ranger, including 10x Genomics tools and third-party tool analysis.
Guidance on next steps after running Cell Ranger, including 10x Genomics tools and third-party tool analysis.
Publication Highlight: Benchmarking scRNA-seq Batch Correction Methods
Publication Highlight: Benchmarking scRNA-seq Batch Correction Methods
Tran et al. performed comprehensive comparisons of 14 batch correction methods. Different methods performed best for 10x Genomics datasets with different characteristics.
Tran et al. performed comprehensive comparisons of 14 batch correction methods. Different methods performed best for 10x Genomics datasets with different characteristics.
10 Tips for Biologists Learning Bioinformatics
10 Tips for Biologists Learning Bioinformatics
Getting started in bioinformatics can feel overwhelming. In this article, you will find some tips and tricks that have helped us on this journey.
Getting started in bioinformatics can feel overwhelming. In this article, you will find some tips and tricks that have helped us on this journey.